ViSeCut
ViSeCut is a medical open-source computed tomography (CT) image viewing-, image processing- and image segmentation software that processes the read two-dimensional computed tomography images (in DICOM format) by various imaging processes, methods and algorithms (volume rendering, watershed segmentation and region-growing segmentation) and represents the desired results. ViSeCut is implemented on Linux (Fedora 10) with programming language C++ and can be used and further expanded by integrating certain medical open-source/cross-platform libraries on Windows, Mac OS X, etc.
Goal
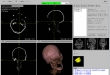
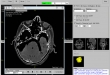
The goal of this project was, implementing an open-source computer program that in the first step reads DICOM formatted 2D CT images, then represents images in 2D axial, sagittal, coronal, and 3D multiplane view. In the second step, the loaded images are visualized and processed in different views at different windows, such as zoom in/out, changing of the position of a view in the window, windowing with Hounsfield-scale, scrolling of images, changing of opacity, changing of color, point selection, screenshot, rotating of image, list specific information from the patient and CT images, such as patient name, number of pictures, picture size, etc.
In the third step, the 2D images should be visualized in a larger window as Cine Mode and scrolled forward and backward for all views automatically. The Cine Mode should be used for enlarged representations of 2D images and 3D models.
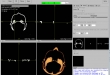
In the next step, 2D images should be converted by specific algorithms (volume rendering, watershed segmentation and region-growing segmentation) in a 3D Model/Object. Before segmentation the program should be able the fact that the user selects itself on 2D images a region (cutting area) and afterwards segmentation algorithm should be selected, thus the processing time is reduced and the algorithm will save time.
Finally segmented 3D Model should be stored as a file in vtk-data format, so that it canb e processed or regarded in another medical computer programs.
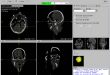
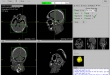
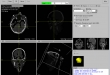
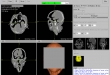
Screenshots
Features
- Easy and user-friendly handling
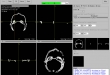
- Visualization of the loaded 2D-images as axial, sagittal, coronal, multiplane, and 3D view (DICOM representation)
- Image viewing for 2D images by using of mouse buttons: zoom in/out, image positioning of all views at separate windows, windowing with Hounsfield scale, rotating of 3D model, point selection of 2D images to display all the views of the same 2D slice image, region selection of 2D images for the cutting function, scrolling
- Image processing for 2D Images: Changing of opacity.
- Image processing for 3D Model: Rotating and changing the color of model
- List of specific image and patient information
- Input field for specific values for Hounsfield scale, marching cubes volume rendering, watershed segmentation and region growing segmentation
- Orientation with labels and a 3D orientation-model by the processing and viewing of 2D images and 3D models
- Cine Mode for forward and backward scrolling of 2D images automatically
- Larger representation of all views in a separate window
- Screenshot for all views
- Exporting of 3D models as vtk-data file
- Live log function for user actions and program events
- Help window
- Loading the CT images with different sizes (512×512, 256×256, etc.)
- Preview function before loading the CT images
Y. Özbek, W. Freysinger
04.03.2012